Introduction
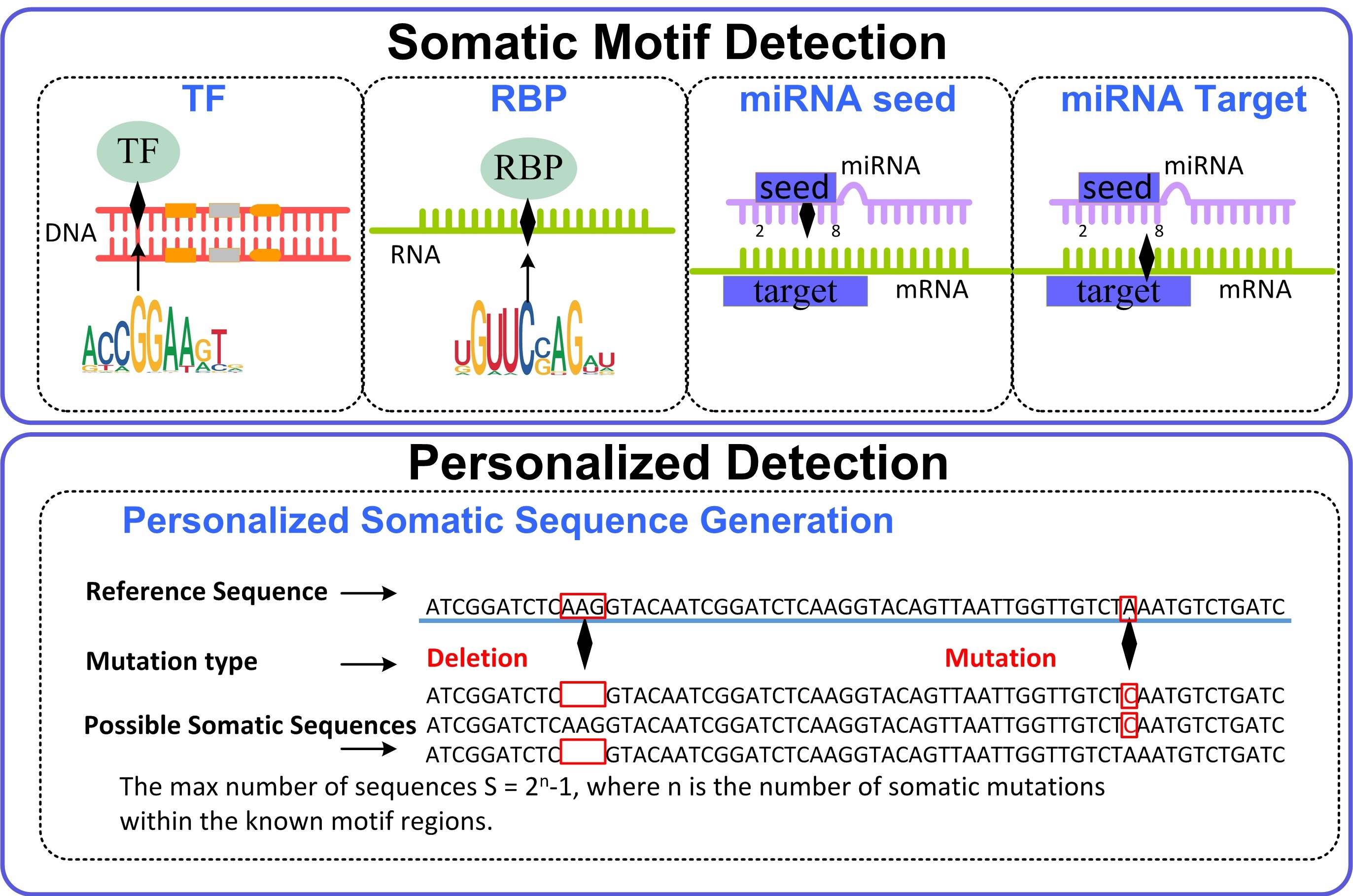
Binding motifs for transcription factors, RNA binding proteins and microRNAs, etc. are vital for proper gene transcription. Sequence altering mechanisms can cause either gain or loss of important binding motifs and thereby lead to severe consequences such as tumorigenesis. Efficient identification of somatic alterations in DNA motifs can provide important insights into the mechanism underlying aberrant gene transcription in human tumors, and thus offer novel cancer therapeutic hypotheses. Altered binding sequences can arise from single nucleotide mutations, insertion, deletion, RNA editing, or single nucleotide polymorphism (SNP). In this database, we successfully identified millions of altered binding motifs, including those for important transcription factor (TF), RNA-binding proteins, and miRNAs seeds and miRNA-mRNA 3’UTR target motifs.
Citation
Framework
Update
Release 2.0 (2020-08-30)
Release 1.0 (2020-07-21)