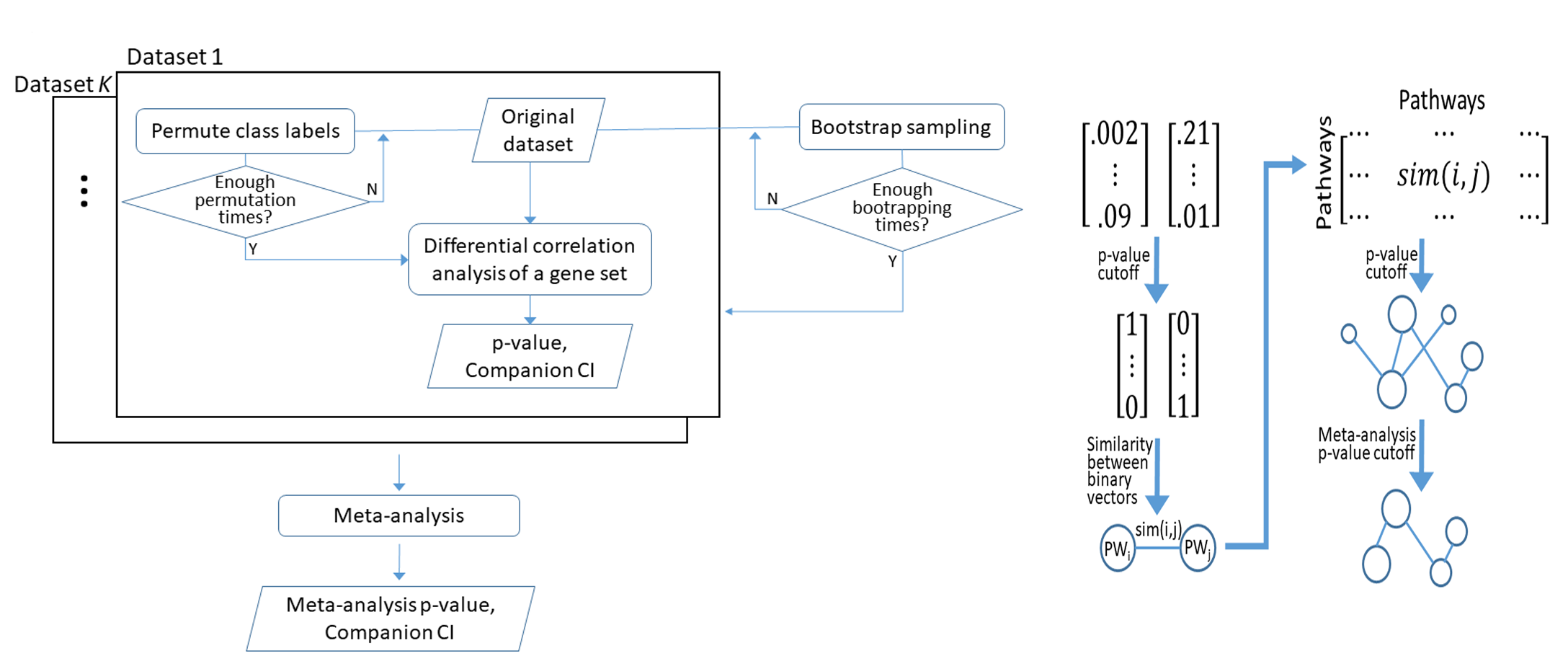
Meta Gene Set Co-Expression Analysis (MetaGSCA) is an analytical tool developed to systematically assess the co-expression of a specified gene set by aggregating evidence across studies. A nonparametric approach that accounts for the gene-gene correlation structure was used to test whether the gene set is differentially co-expresssed between two comparative conditions, from which a permutation test p-value was computed. Bootstrapping was used to construct a confidence interval of the permutation p-value. A meta-analysis of permutation p-values from individual studies is then performed with one of two options: random-intercept logistic regression model or the inverse variance method, yielding a conclusive result over a number of individual studies. In addition, a pathway crosstalk network can be delineated to represent pathways’ similar profiles of correlation disturbance across the aligned datasets.