Introduction
Binding motifs for transcription factors, RNA binding proteins and microRNAs, etc. are vital for proper gene transcription. Sequence altering mechanisms can cause either gain or loss of important binding motifs and thereby lead to severe consequences such as tumorigenesis. Efficient identification of somatic alterations in DNA motifs can provide important insights into the mechanism underlying aberrant gene transcription in human tumors, and thus offer novel cancer therapeutic hypotheses. Altered binding sequences can arise from single nucleotide mutations, insertion, deletion, RNA editing, or single nucleotide polymorphism (SNP). The SBSA, as an easy online tool for annotating and identifying altered binding motifs, supports personalized genome approach, customizable motif regions, and a wide selection of species.
Framework
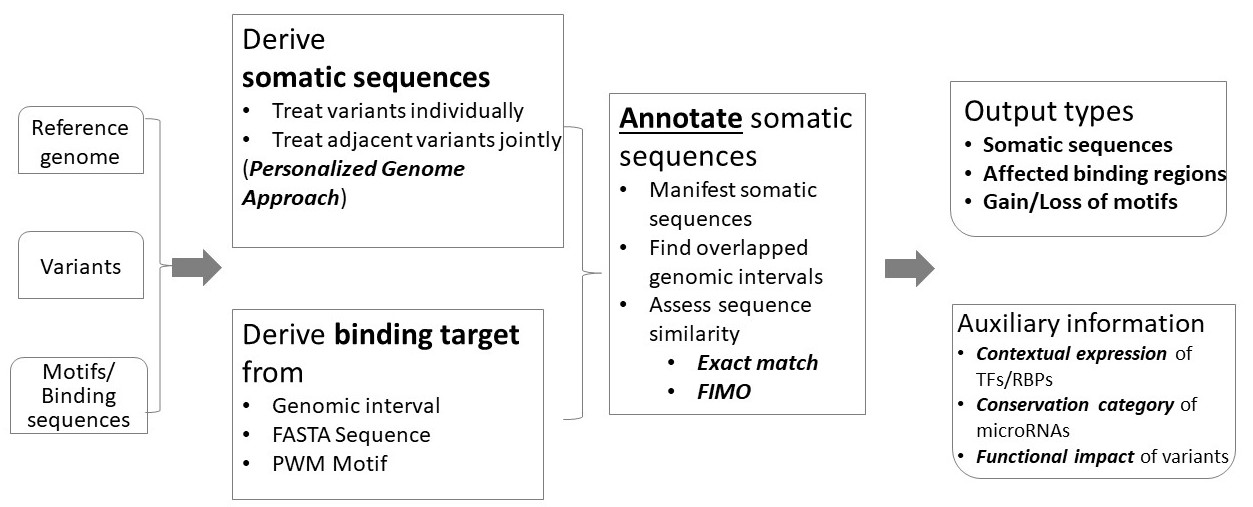
Somatic Binding Sequence Annotator (SBSA) annotates genomic variants with respect to binding motifs of molecule regulators. Of the three inputs, variant specification file is mandatory, while the other two can be preloaded datasets. SBSA supports 26 reference genomes and harbors abundant motifs/binding sequences for three major types of transcription regulators: TFs, RBPs, and microRNAs. Depending on the type and specification of inputs, SBSA performs a relevant annotation of the input variants and generates a corresponding output. For TF/RBPs, gain or loss of binding motifs is affirmed by either FIMO or an intuitive Exact match method; for microRNA seeds or microRNA binding targets, the affected genomic regions are identified; in the simplest work mode, somatic sequences centered upon the input variants are generated. TF, transcription factor; RBP, RNA-binding protein; FIMO, the bioinformatics algorithm Find Individual Motif Occurrences; FASTA, the sequence file format for Fast Adaptive Shrinkage/Thresholding Algorithm; PWM, Position-Weight Matrix.
Update
more...
Release 2.2 Added “Fine-mapping results” of CAVIAR, CaVEMaN, and DAP-G, Contextual Expression of TF/RBP genes in GTEx Analysis V8, conservation and annotation quality of miRNA, and 20 scores to evaluate variant effects. Updated the Help section.
Added the function of scaning motif. Added 11 preprocessed TF motif databases. Changed the output format for sequence. And allowed any genome.
Added 19307 SARS genomes. And added the help section.
The first version of SBSA Tool.
Citation
Limin Jiang, Fei Guo, Jijun Tang, Hui Yu, Scott Ness, Mingrui Duan, Peng Mao, Ying-Yong Zhao, Yan Guo, SBSA: an online service for somatic binding sequence annotation, Nucleic Acids Research, Volume 50, Issue 1, 11 January 2022, Page e4, https://doi.org/10.1093/nar/gkab877