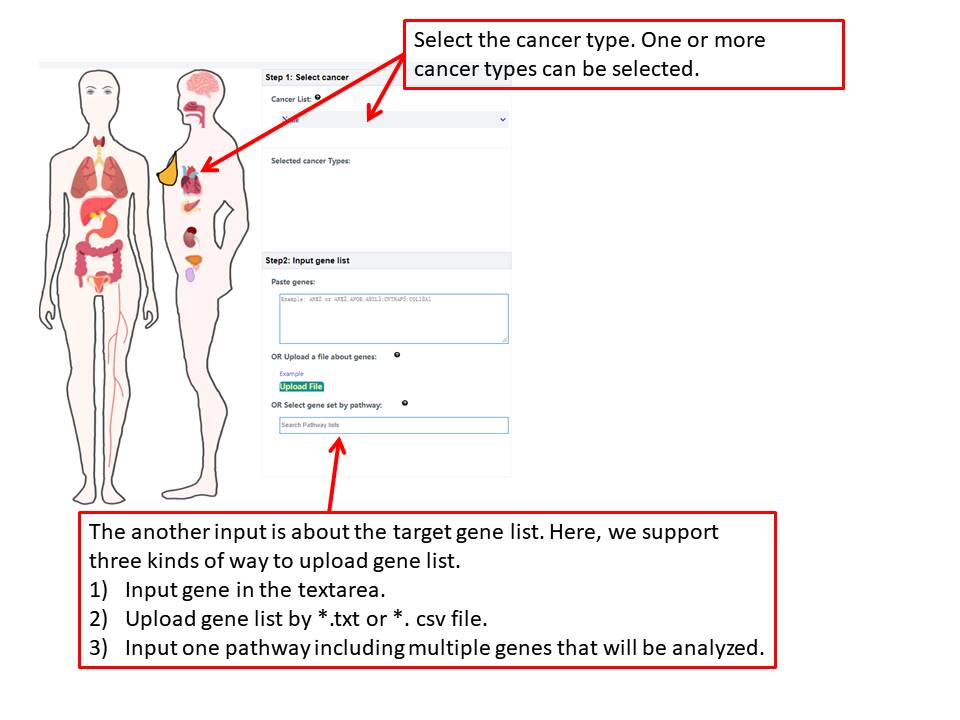
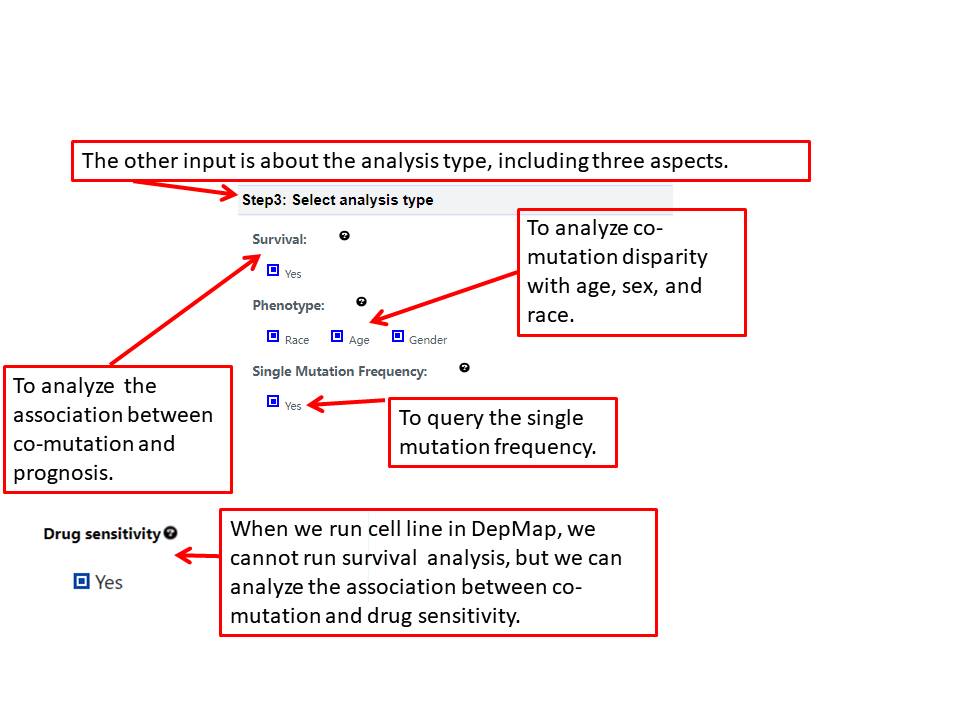
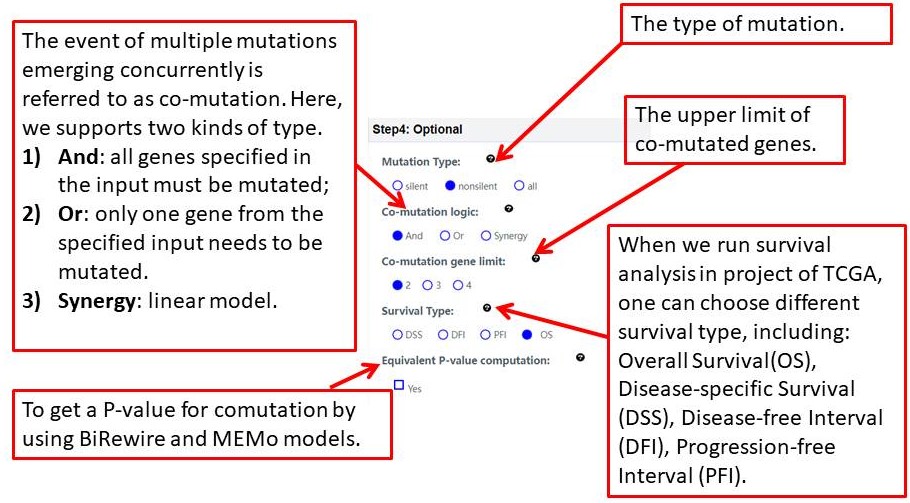
This section will explain the all parameters and results in CoMutDB database.
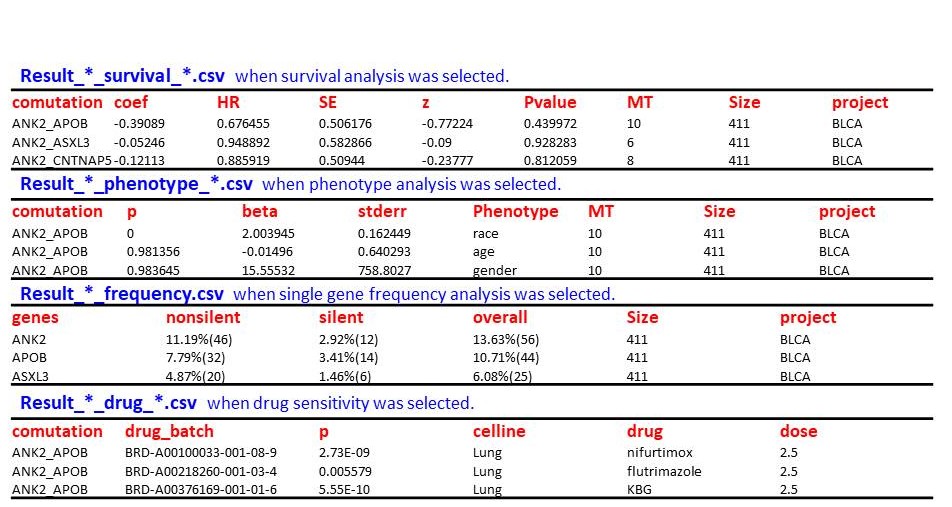
Header | Meaning |
Alt | Altered nucleotide |
beta | The beta is coefficient estimate that indicate the average change in the log odds of the response variable associated with a one unit increase in each predictor variable |
celline | The name of cell line |
comutation | Gene names in co-mutation pair |
coef | The log hazard ratios |
comut_P | The equivalent p value for co-mutation by using BiRewire R package. It can be found in the Survival/Phenotype/Drug sensitivity result tables if you want to evaluate statistical significant of co-mutation. |
Chr | Chromosome where a variant is located |
class | The type of mutation |
drug | The name of drug |
drug_batch | |
dose | The dose of drug |
Frequency | The frequency of single mutation |
Fgene1 | The frequence of the first gene |
Fgene2 | The frequence of the second gene |
genes | The gene name |
Gene | Symbol of the host gene whose body encloses or approximates the variant |
HR | The hazard ratios |
MT | The number of samples with co-mutation |
nonsilent | The frequency of nonsilent mutation |
overall | The frequency of all mutation |
Phenotype | Phenotype type |
Pvalue | The propability that co-mutation don't associate with prognosis |
project | The cancer project name or cell line name |
p | The propability that co-mutation don't associate with phenotype |
Pos | Chromosome coordinate position of a variant |
Ref | Reference nucleotide |
silent | The frequency of silent mutation |
SE | The standard error |
Size | The number of samples |
stderr | Standard error |
z | Z-score |