This section will explain the all parameters and results in MutMix database.
Index
MutMix Modules
Notes on query output
Notes on query input
Table headers
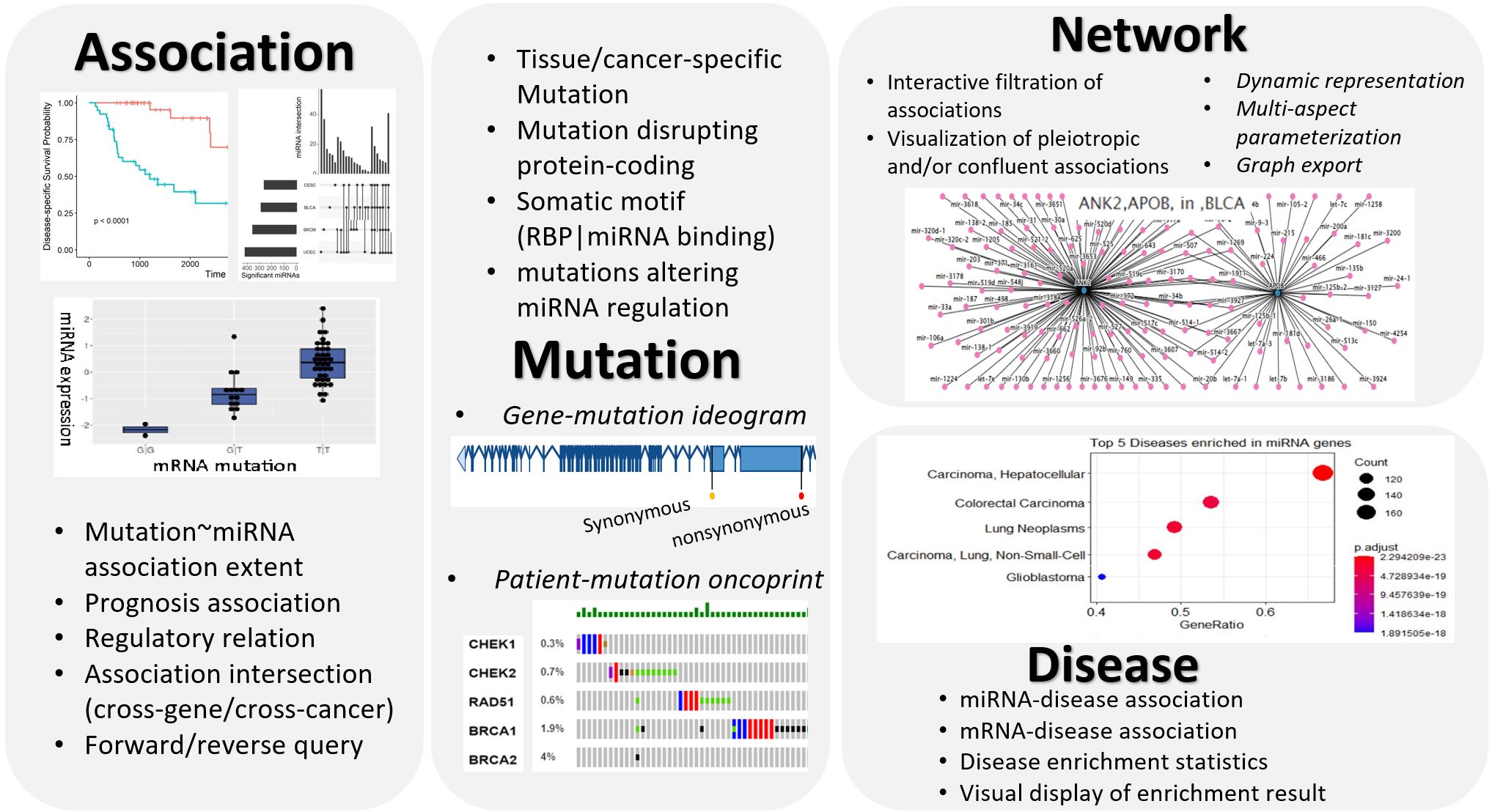
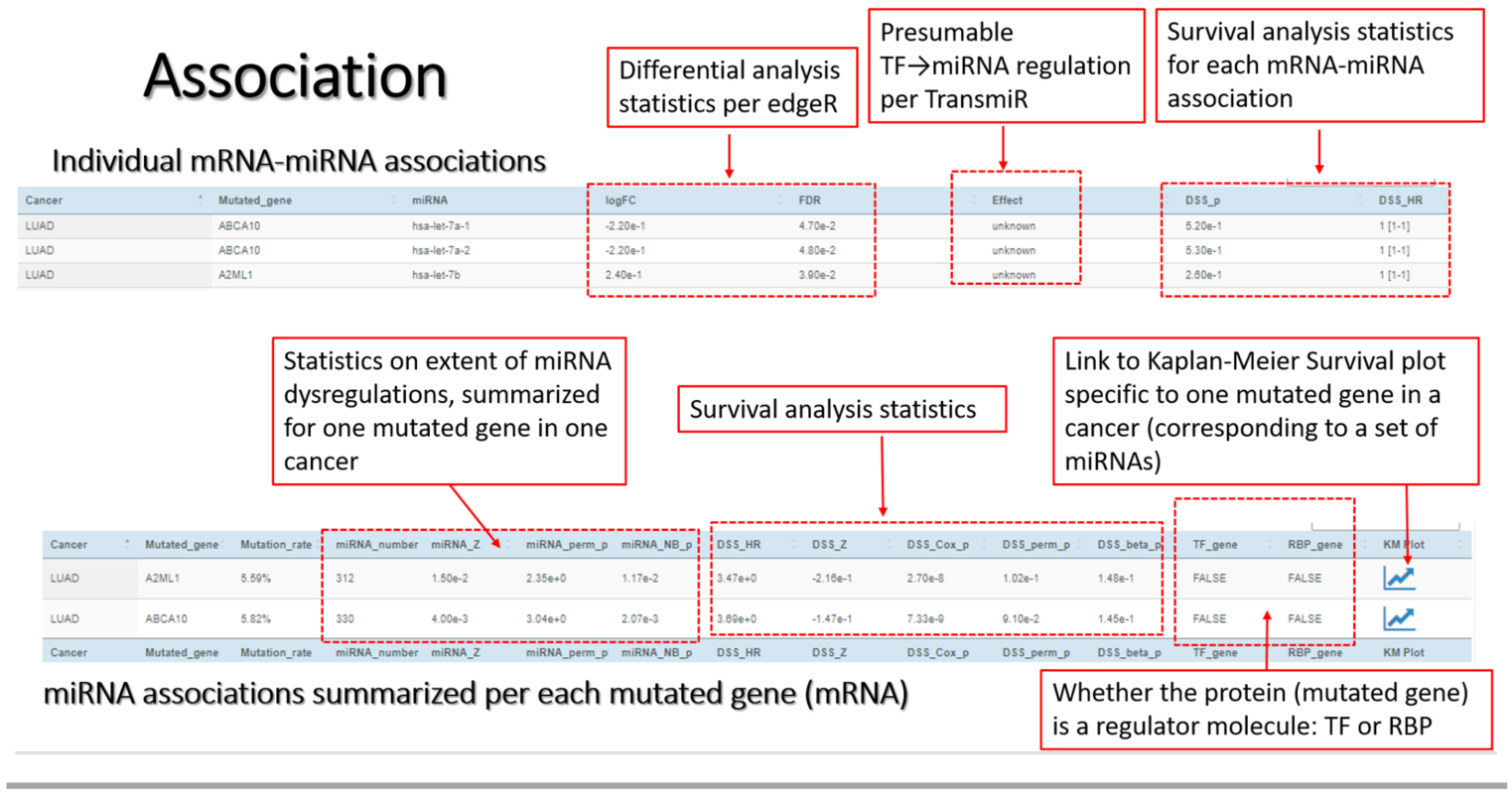
*Association* Using rigorous statistical methods, MutMix infers mutation-miRNA association based on genome sequencing data of mRNA genes and RNA-Seq data of miRNAs. Presumable TF→miRNA relationships are evidenced with TransmiR. A surrogate CGES expression score is summarized for all miRNAs associated with the common mutated gene, and its prognosis predictability is assessed with respect to Disease-Specific Survival or Overall Survival. Intersection of mRNA-miRNA associations can be interrogated in either a cross-gene or a cross-cancer manner.
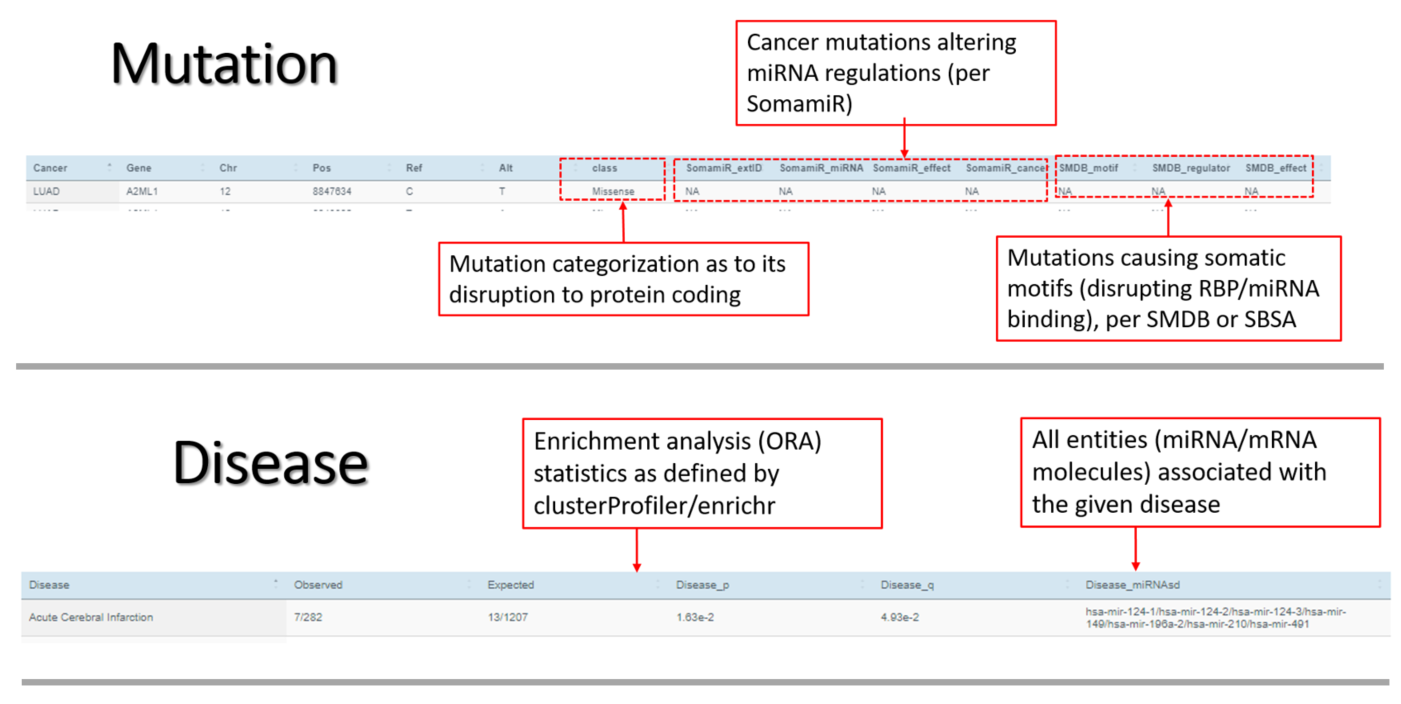
*Mutation* Mutations are tabulated and visually displayed. The tabular representation reveals the chromosome coordinates, host gene name, protein-coding disruption, tissue-specificity, and regulatory alternations (RBP/miRNA binding). A gene-body ideogram indicates the intron-exon linear structure marked with categorized mutations. A patient-mutation oncoprint resorts to the heatmap concept to visualize the occurrence of categorized mutations in diverse genes across individual patients.
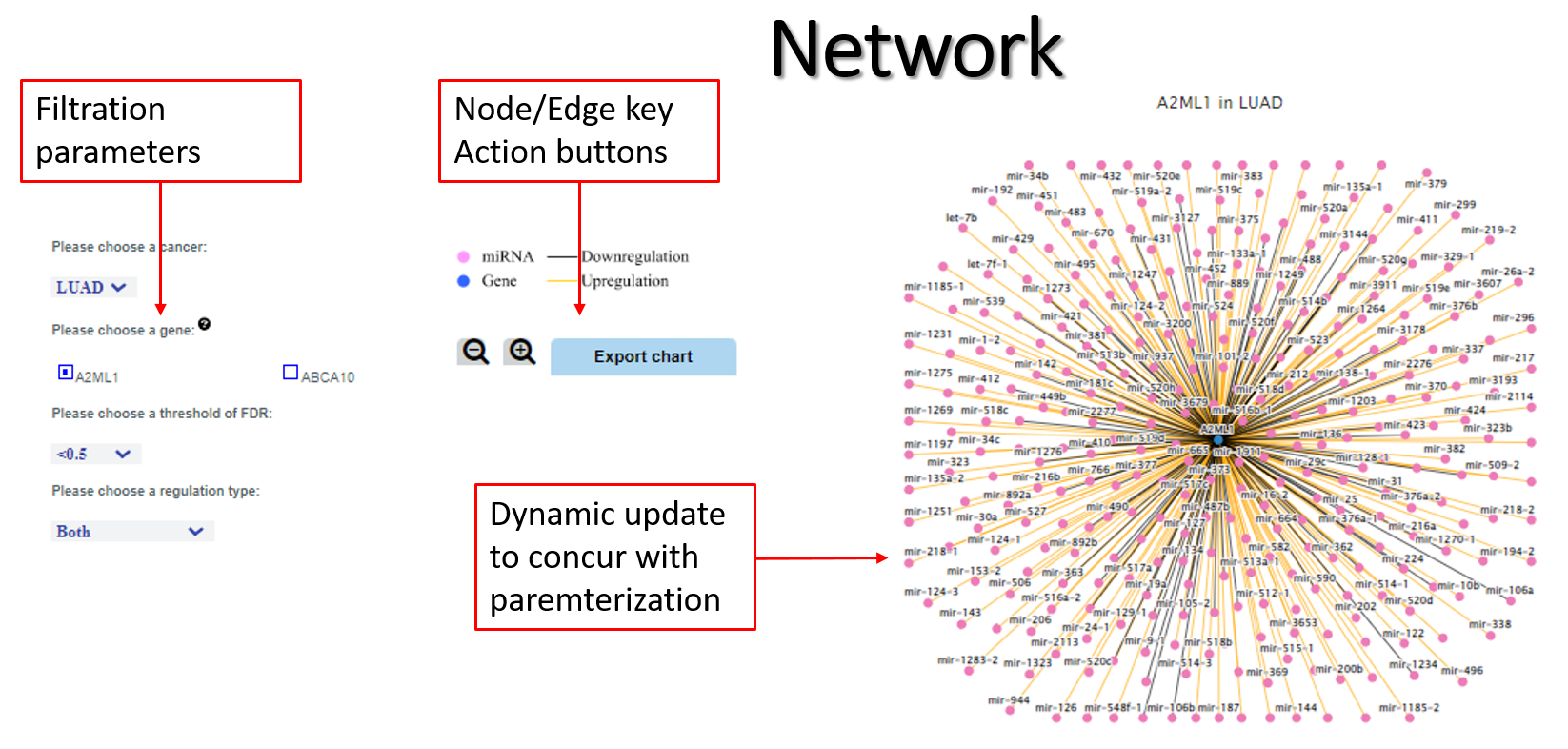
*Network* mRNA-miRNA associations are visually represented in a network, which can be updated and exported to reflect the user’s interactive parametrization. Pleiotropic and/or confluent associations are more notable in the network representation than in the tabular form. The network can be centered on select protein-coding genes (forward query) or select miRNAs (reverse query).
*Disease* Disease associations for a set of miRNAs/mRNAs undergo an enrichment analysis (over-representation analysis), and the most prominent diseases are returned in a ranked list. Visualization of the top-ranking enriched diseases is provided.
| Header | Meaning |
| Alt | Alternative allele of a genomic variant |
| Cancer | The cancer in which an association was inferred |
| Chr | Chromosome |
| Class | Mutation class (synonymous, nonsynomous, stopgain, etc) |
| Disease | The disease associated with miRNA(s) |
| Disease_miRNAs | miRNAs associated with a disease |
| Disease_p | p-value of disease enrichment among a set of miRNAs |
| Disease_q | q-value of disease enrichment among a set of miRNAs |
| DSS_beta_p | p-value of miRNA-set's Disease-Specific Survival (DSS) prognosis, inferred via Beta distribution of permutation data |
| DSS_Cox_p | p-value of miRNA-set's DSS prognosis, inferred via Cox proportional-hazards model |
| DSS_HR | Hazard Ratio (HR) of DSS prognosis association, inferred via Cox proportional-hazards model |
| DSS_p | p-value of single miRNA's DSS prognosis, inferred via Cox proportional-hazards model |
| DSS_perm_p | p-value of miRNA-set's DSS prognosis, inferred via permutation data |
| DSS_Z | Z-score of miRNA-set's DSS prognosis, inferred via permutation data |
| Effect | Putative regulatory effect between a TF and a miRNA, per TransmiR |
| Expected | Expected proportion of disease-associated miRNAs in a miRNA set |
| FDR | False Discovery Rate, in miRNA dysregulation statistical test (per edgeR) |
| KM Plot | Kaplan-Meier survival plot |
| LogFC | Logged fold change, in miRNA dysregulation statistical test (per edgeR) |
| miRNA | microRNA name |
| miRNA_NB_p | p-value for mutation-associated miRNA dysregulation extensity, inferred via Negative Binomial distribution |
| miRNA_number | number of dysregulated miRNAs associated with the mutated gene |
| miRNA_perm_p | p-value for mutation-associated miRNA dysregulation extensity, inferred via permutation data |
| miRNA_Z | Z-score for mutation-associated miRNA dysregulation extensity, inferred via permutation data |
| Mutated_gene | The mutated gene (gene symbol) |
| Mutation_rate | Proportion of patients in a cancer-type cohort who carry a mutation in the concerned gene |
| Observed | Observed proportion of disease-associated miRNAs in a miRNA set |
| OS_beta_p | p-value of miRNA-set's Overall Survival (OS) prognosis, inferred via Beta distribution of permutation data |
| OS_Cox_p | p-value of miRNA-set's OS prognosis, inferred via Cox proportional-hazards model |
| OS_HR | Hazard Ratio (HR) of OS prognosis association, inferred via Cox proportional-hazards model |
| OS_p | p-value of single miRNA's OS prognosis, inferred via Cox proportional-hazards model |
| OS_perm_p | p-value of miRNA-set's OS prognosis, inferred via permutation data |
| OS_Z | Z-score of miRNA-set's OS prognosis, inferred via permutation data |
| Pos | Chromosome coordinate position of a variant |
| RBP_gene | Whether the mutated gene is a RNA-binding protein (RBP) |
| Ref | Reference allele of a genomic variant |
| SMDB_effect | The type of somatic motif per the implicated regulator, also the sector of SMDB |
| SMDB_motif | Mutation-caused somatic motif (new form), per SMDB |
| SMDB_regulator | The regulator (RBP, miRNA seed, or miRNA target) implicated with the somatic motif, per SMDB |
| SomamiR_cancer | The related cancer type, per SomamiR |
| SomamiR_effect | The effect of cancer-related somatic mutation, per SomamiR |
| SomamiR_extID | The external ID of the implicated regulator of the somatic mutation, per SomamiR |
| SomamiR_miRNA | The miRNA implicated with the somatic mutation, per SomamiR |
| TF_gene | Whether the mutated gene is a Transcription Factor (TF) |